Transformers are a significant trend in computer vision, and I recently provided an overview of some remarkable advancements. In this instance, I will use my re-implementation of a transformer-based model for 3D segmentation. Specifically, I will be utilizing the UNETR transformer and analyzing its performance in comparison to a classical UNET. The notebook is available.
UNETR is the first successful transformer architecture for 3D medical image segmentation. I will attempt to match the results of a UNET model on the BRATS dataset, containing 3D MRI brain images. I will be training the UNETR transformer for this tutorial.
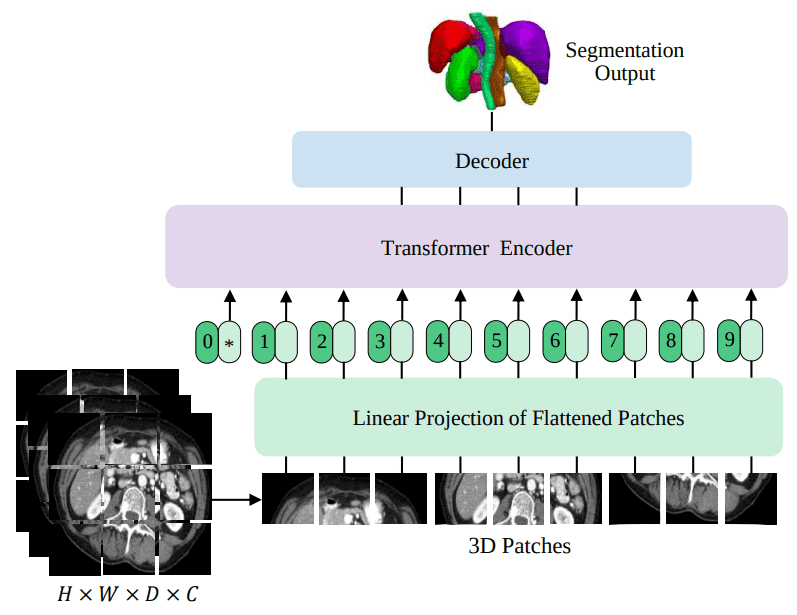
Source: UNETR: Transformers for 3D Medical Image Segmentation, Hatamizadeh et al.
My model was tested using an existing tutorial on a 3D MRI segmentation dataset. Credit goes to the amazing open-source library of Nvidia called MONAI for providing the initial tutorial that I modified for educational purposes. If you are interested in medical imaging, be sure to check out this incredible library and its tutorials.
The BRATS dataset is a multi-modal large-scale 3D imaging dataset, containing 4 3D volumes of MRI images captured under different modalities and setups. It is important to note that only the tumor is annotated in the dataset, making things like segmentation more difficult. A breakdown of the tumor categories, and types of segmentations and labels is provided using a teaser image from the BRATS completion website.
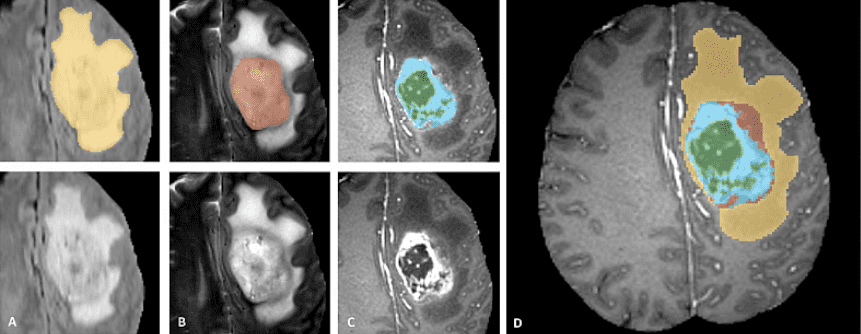
BRATS dataset
Editing the dataset from the medical imaging decathlon competition becomes trivial with MONAI.
Data loading with MONAI and transformations
Using the DecathlonDataset class of the MONAI library, any of the 10 available datasets can be loaded from their website. For this tutorial, the Task01_BrainTumour dataset is used. The transformation pipeline plays a crucial role in this process, as 3D images require extensive adjustments and MONAI provides functions to make this easier.
A pipeline showcasing the transformation is visualized using a sample of the train data below: (Image Included)
The segmentations in the pipeline are adjusted to ensure a smoother version of the original labels. The class ConvertToMultiChannelBasedOnBratsClassesd is used for this purpose.
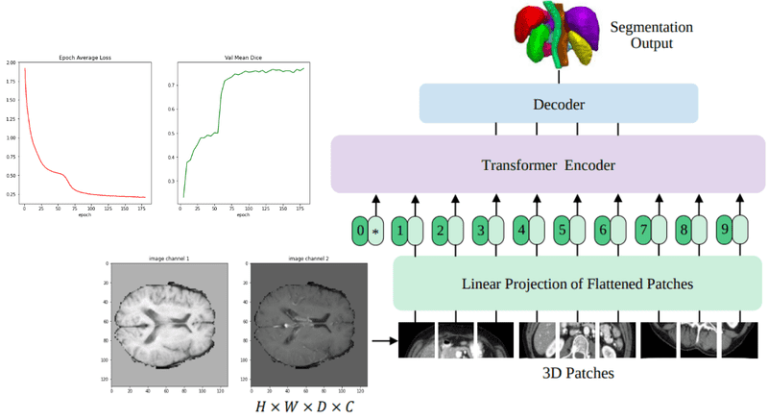
The UNETR architecture
At this point, the model architecture that integrates transformers into the UNET architecture is exemplified. An image illustrating the model architecture is provided (Image Included).
I also compared my implementation with MONAI’s UNETR implementation to test for performance and gain insights on architectural changes. Notably, significant details are often excluded in architectural images, and it’s recommended to check the implementation code for complete information.
An example of how to initialize the model is shown below:
$ pip install self-attention-cv==1.2.3
Finally, the model’s performance is analyzed by comparing the Dice coefficient while training. The performance of the UNETR model is tracked through COVID-19 data, kindness of the author’s implementation. It is evident that the most crucial aspect to obtain a good performance, particularly as seen from the DICE coefficient, is the data preprocessing and transformation pipelines. This calls for limited innovation in the medical imaging world for machine learning modeling, focusing more on data processing optimization.
Conclusion and concerns
The author expresses some concerns regarding the performance of transformers in 3D medical imaging, admitting that more advanced methods and other contributions will likely follow. It highlights the crucial role of data preprocessing and transformation pipelines in achieving good performance, raising concerns about new papers and architectural claims not being fair. The author also appreciates community support for the book “Deep learning in production.”
Deep Learning in Production Book 📖
Learn how to build, train, deploy, scale and maintain deep learning models. Understand ML infrastructure and MLOps using hands-on examples.
* Disclosure: Please note that some of the links above might be affiliate links, and at no additional cost to you, we will earn a commission if you decide to make a purchase after clicking through.